Searching a protein in CoDNaS
There are many ways to find your query protein in CoDNaS database:
Search by PDB code or UniProt id in the home page
The user can write a PDB code or UniProt id in the the field and press the search button.

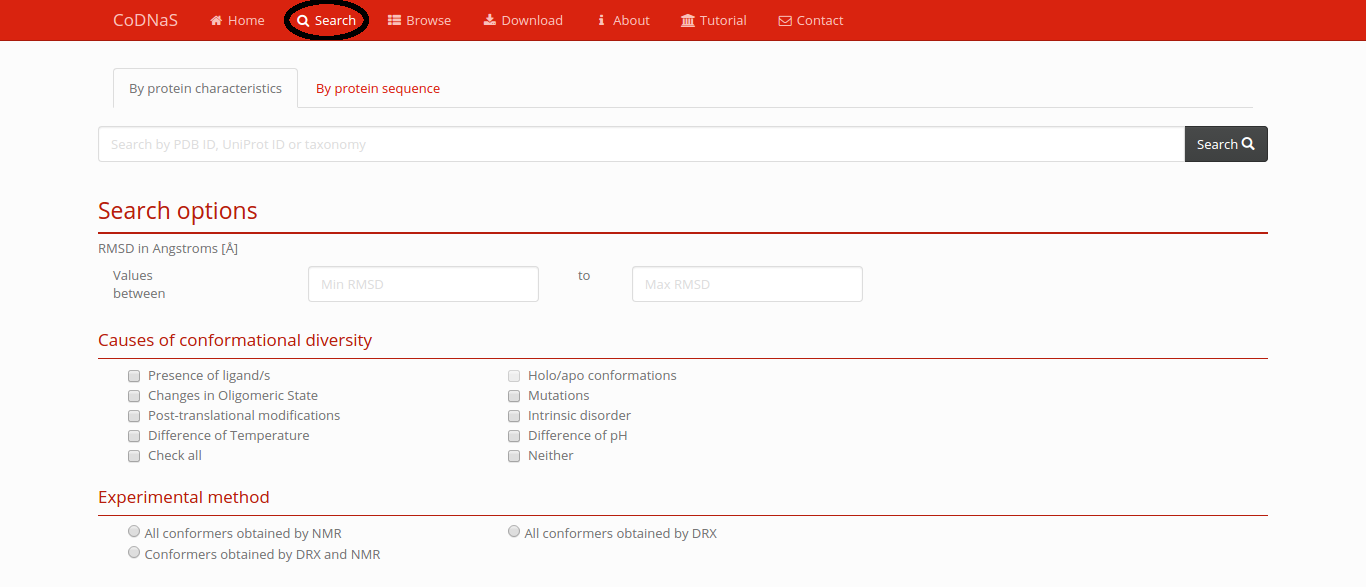
Advanced Search
- By protein characteristics: in this page the user can search a protein query by different criteria.
- PDB code or UniProt ID.
- Extension of conformational diversity of a protein: the user can define a range of RMSD values in order to retrieve protein with a given extension of conformational diversity.
- Causes of conformational diversity: the user can select different causes of conformational diversity in order to retrieve proteins that present pairs of conformers associated to one or more causes.
- Experimental method: the user can filter the experimental method with that have been obtained the conformers of a given protein.
- By protein sequence: in this section the user can put a protein/DNA sequence in fasta format and the server run a BLAST query against all protein sequences in CoDNaS. The retrieved proteins has a BLAST E-value cutoff of 1E-04.
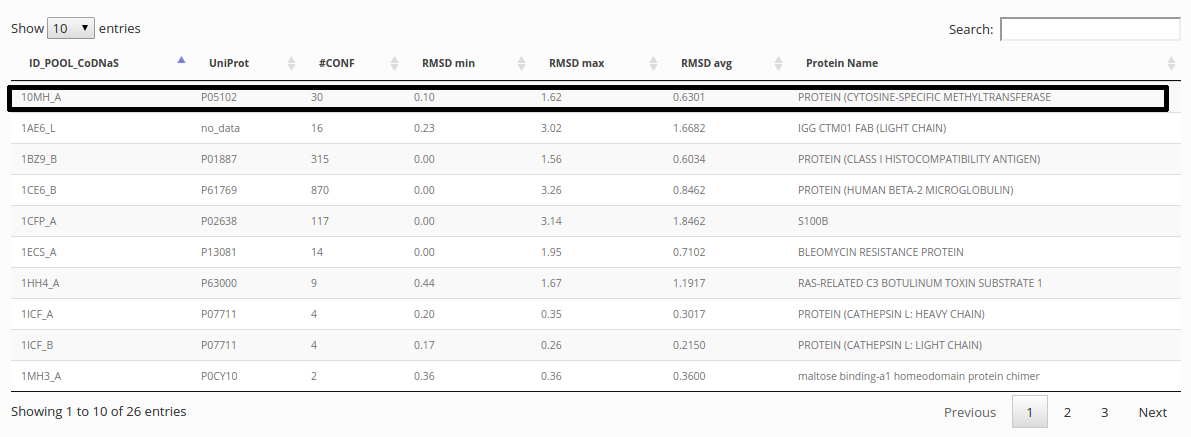
Viewing a protein entry in CoDNaS
After a search, the results page show a table where you can press on a row in order to enter in a protein description page.
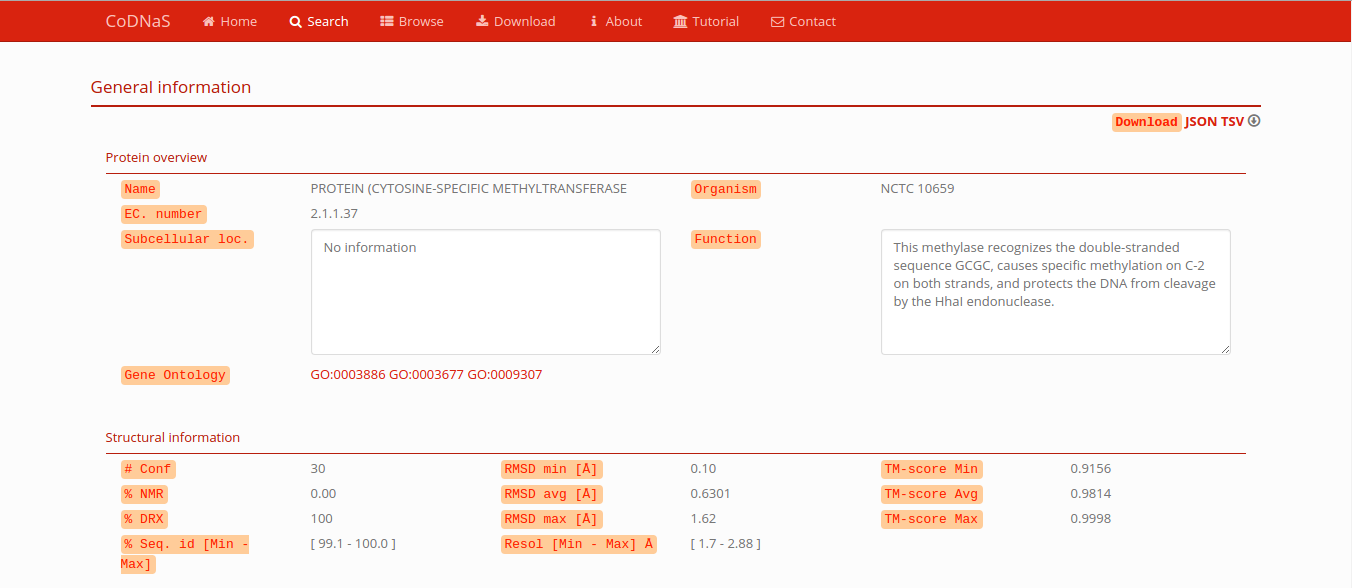
General information of the protein
This first section have biological information about the current protein and general structural information: amount of conformers, percentage of conformers obtained by Nuclear Magnetic Resonance (NMR) of X-ray diffraction (XRD), global sequence identity between conformers, comparison between all conformers (RMSD,TM score, etc), and others. If you have to download this information, press the link JSON (JSON format) or TSV (tab-separated values) in the right corner of this section.
Protein chain conformations
List of all protein structures conformations with specific information: experimental method, resolution (only in X-RAY structures), UniProt ID, presence of ligands, presence of Post-translational modifications and missing residues.
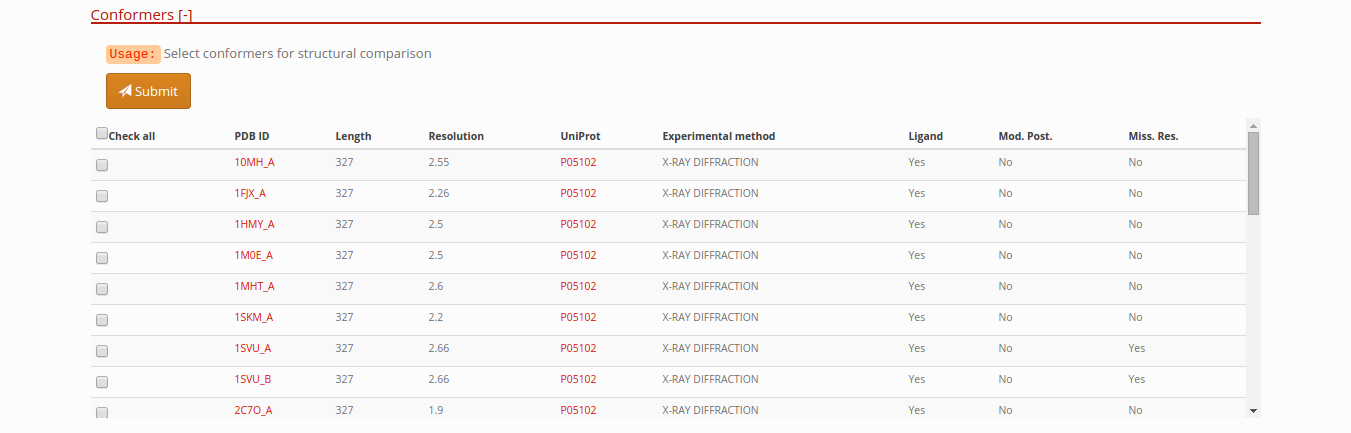
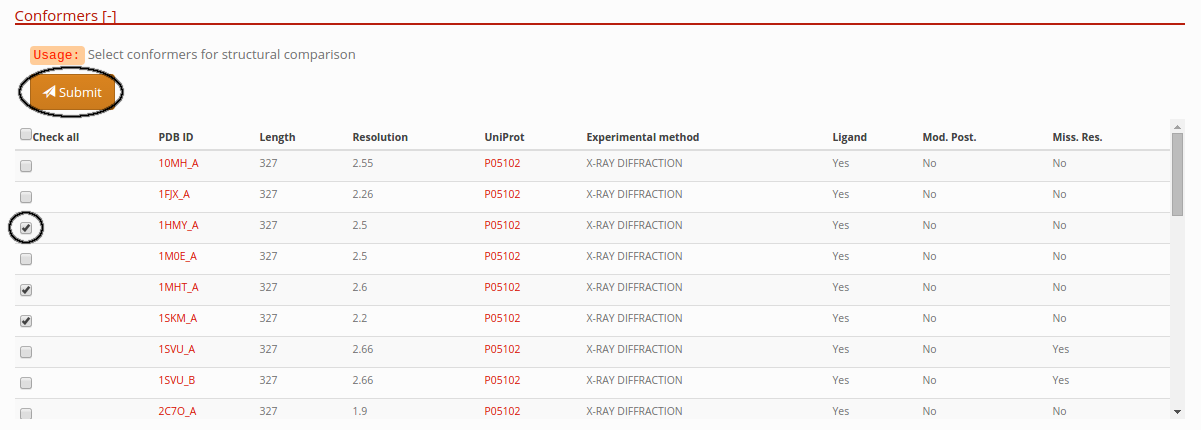
Selecting conformers to compare in a protein entry
In this section of the entry page, you can select different conformations and press the submit button, in order to retrieve all vs all structural alignments comparison. View Comparison of conformers section.
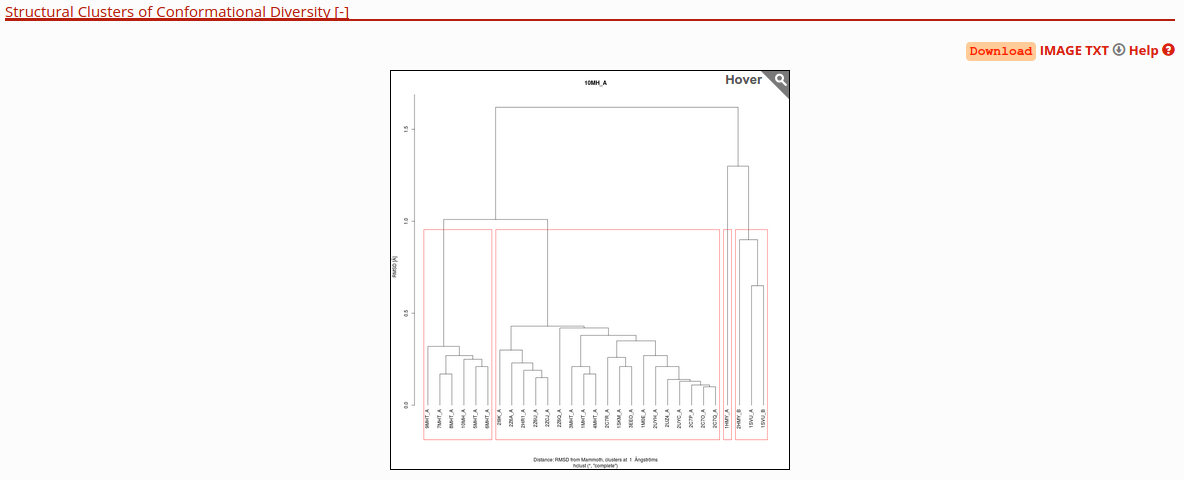
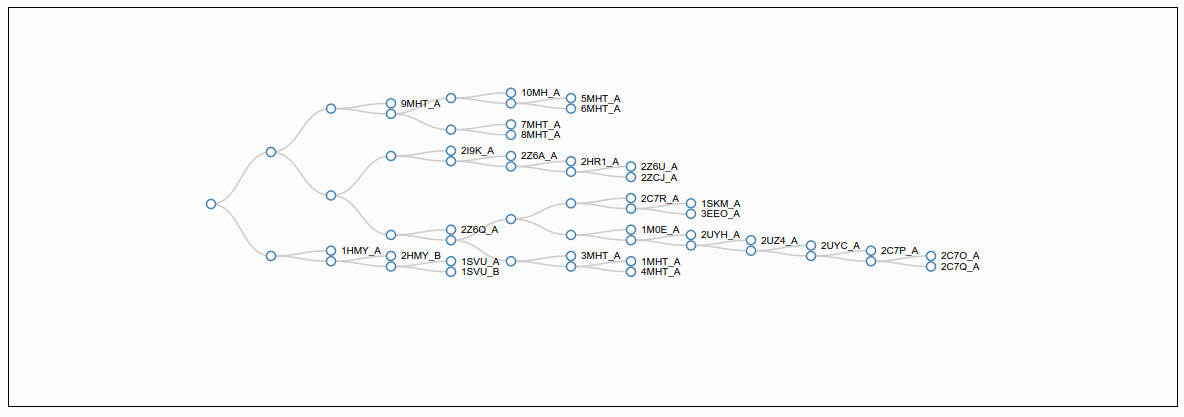
View hierarchical clustering of conformers by RMSD
This section have a dendrogram in the top and a interactive flexible force-directed graph layout that allow the user see the conformers that have similar according to the RMSD value in all vs all comparisons.
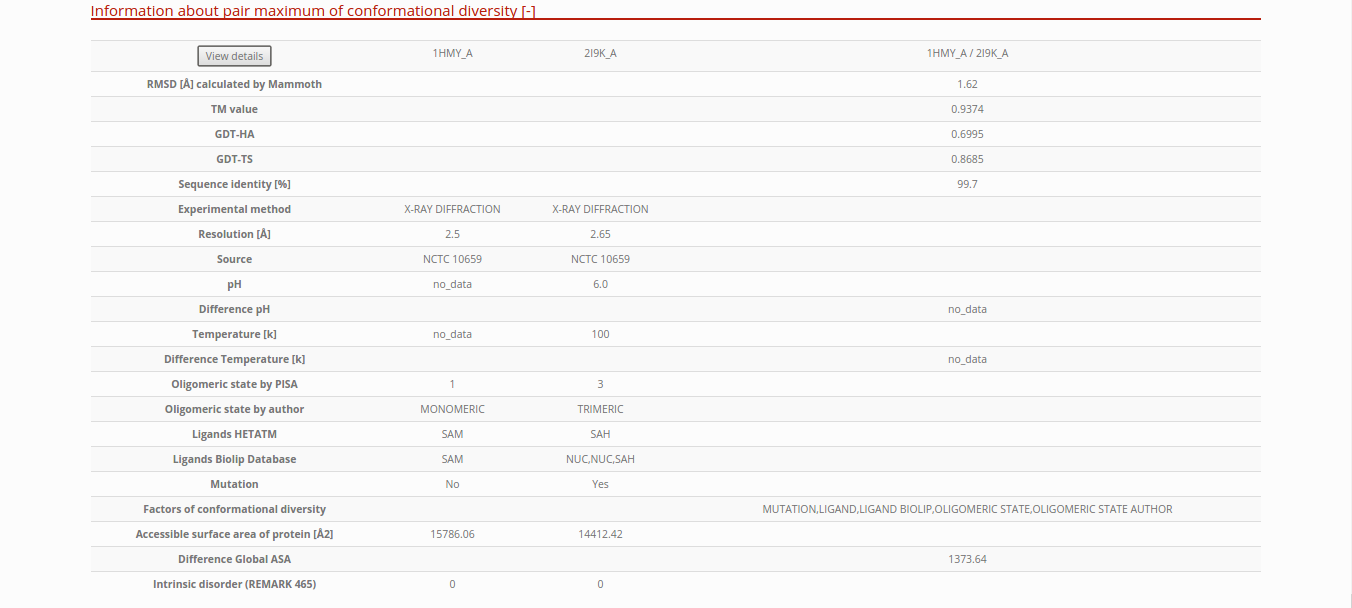
Detailed information about the pair of maximum conformational diversity
In the table, you can see the differences in the experimental conditions and other characteristics about the pair of maximum RMSD in the protein. If you press the “View details” button, you will go to the page of detailed information about pairs of conformers. See more details in the next section of this tutorial.
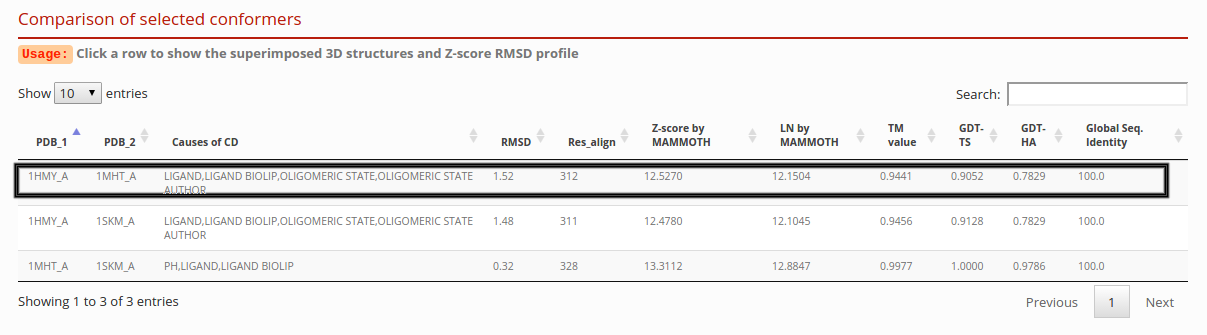
Comparison of conformers
The result table contains all vs all structural information of the selected conformers. You can press any pair of conformers to show a detailed information and view the structural alignment.
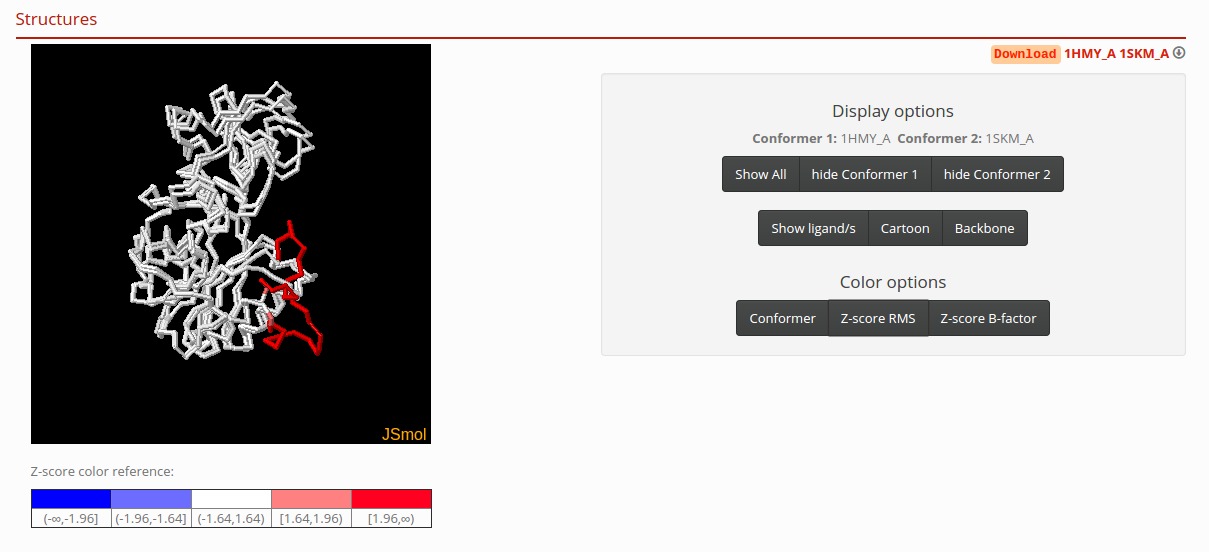
Structures
In this section, you can interact with the Jsmol visualizer. If you click the “Z-score RMS” button in color options, you will see the superposed structures colored according to the extent of C-alpha RMS by position, normalized in Z distribution. In addition, you can click the “Z-score B-factor” button to see superposed structures colored according to the extent of C-alpha B-factors, normalized in Z distribution. If you have to download the superposed structures in a PBD format file, press the links in the right corner of this section.
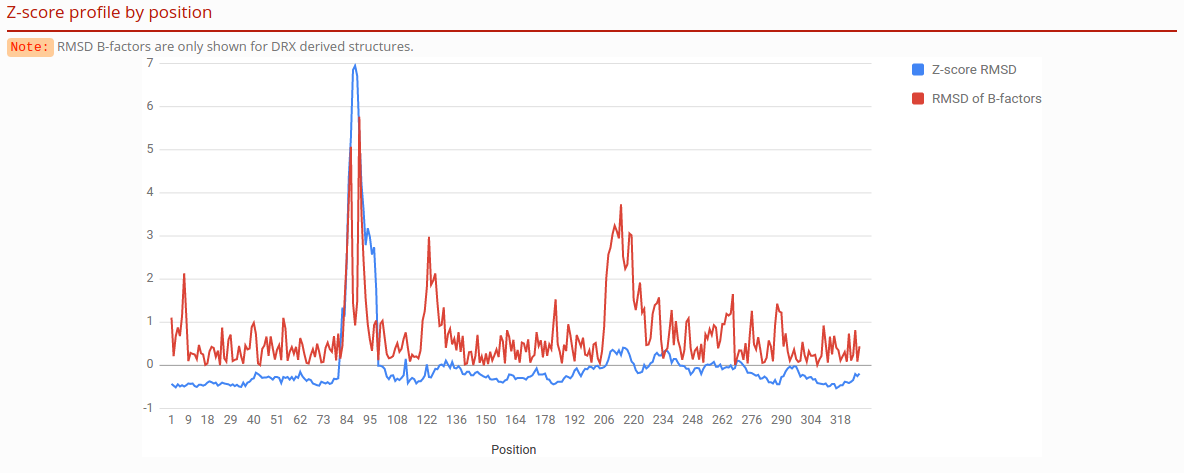
Z-score profile by position
Plot shows C-alpha RMS and C-alpha B-factor RMSD by position, normalized in Z distribution.
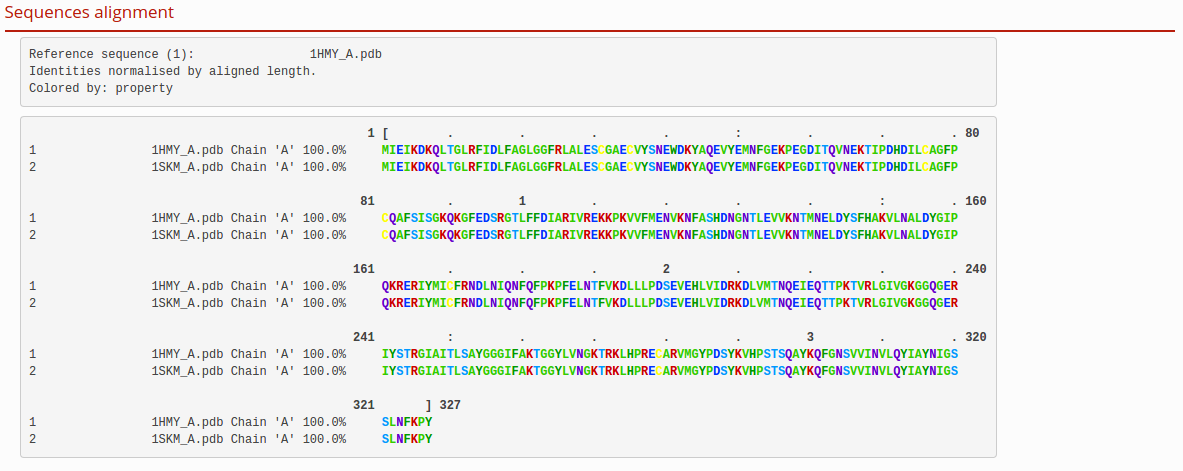
Sequences alignment
The sequence alignment in the next figure is based on the structural superposition realized in the previous step.
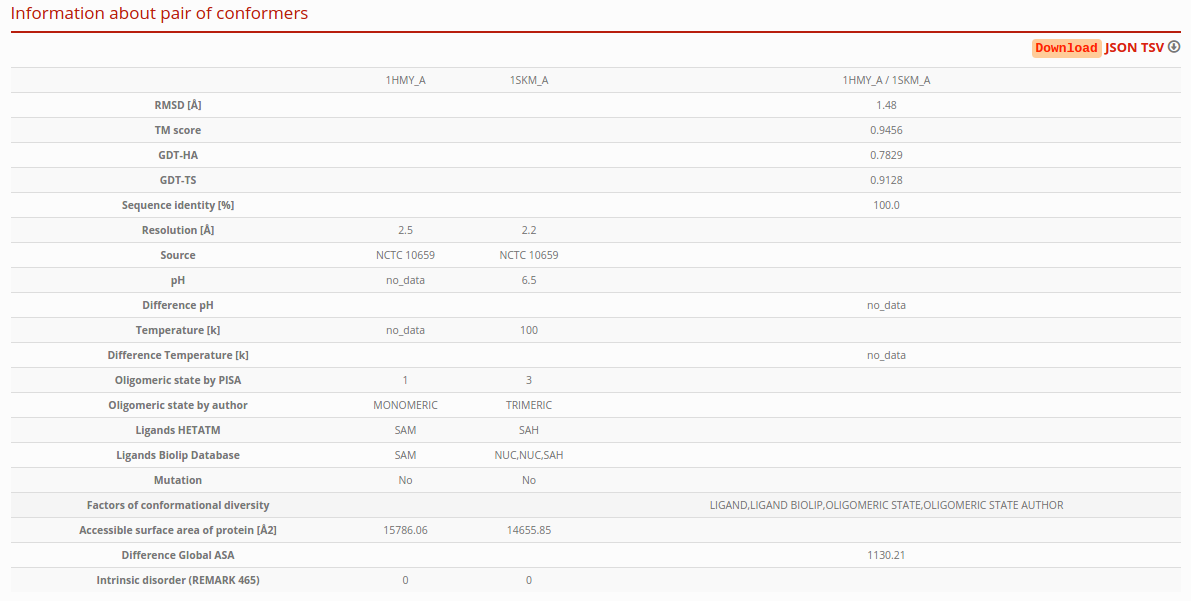
Information about pair of conformers
See details in the section “Detailed information about the pair of maximum conformational diversity”. If you have to download this information, press the link JSON (JSON format) or TSV (tab-separated values) in the right corner of this section.
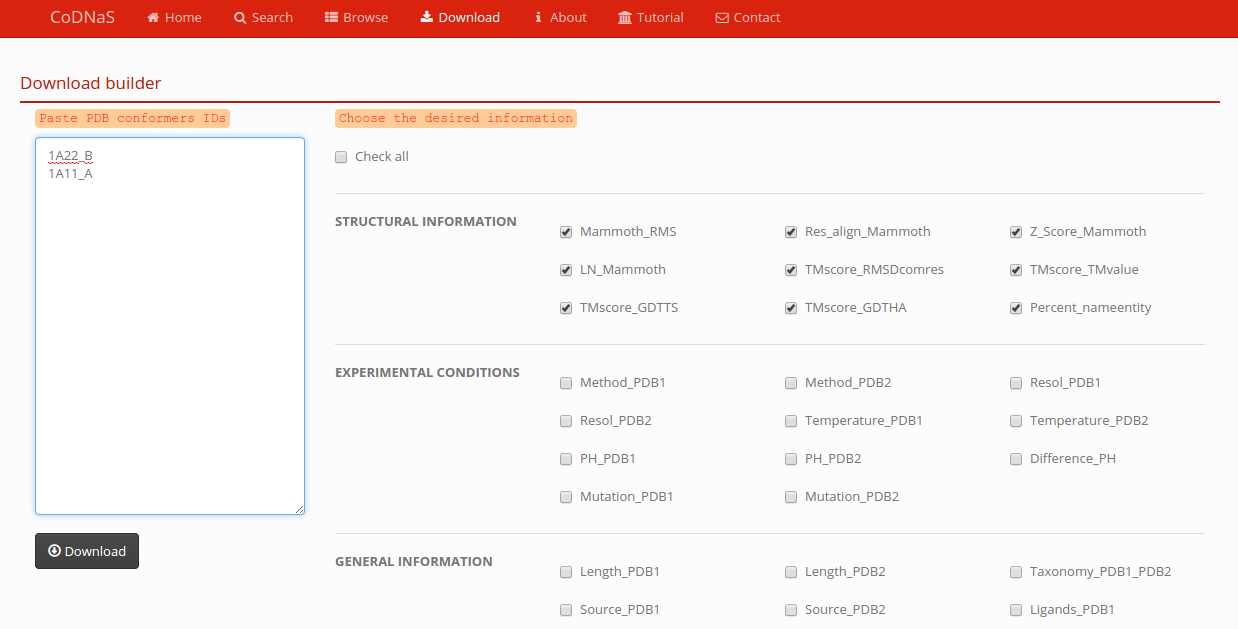
Download
You can build a custom download. Paste conformers PDB codes in the textarea (one id per row), and choose the information you wan't. You will get all the pair comparison for the selecteds PDBs along with the information you choose to download. Finally you get a tab-separated file.
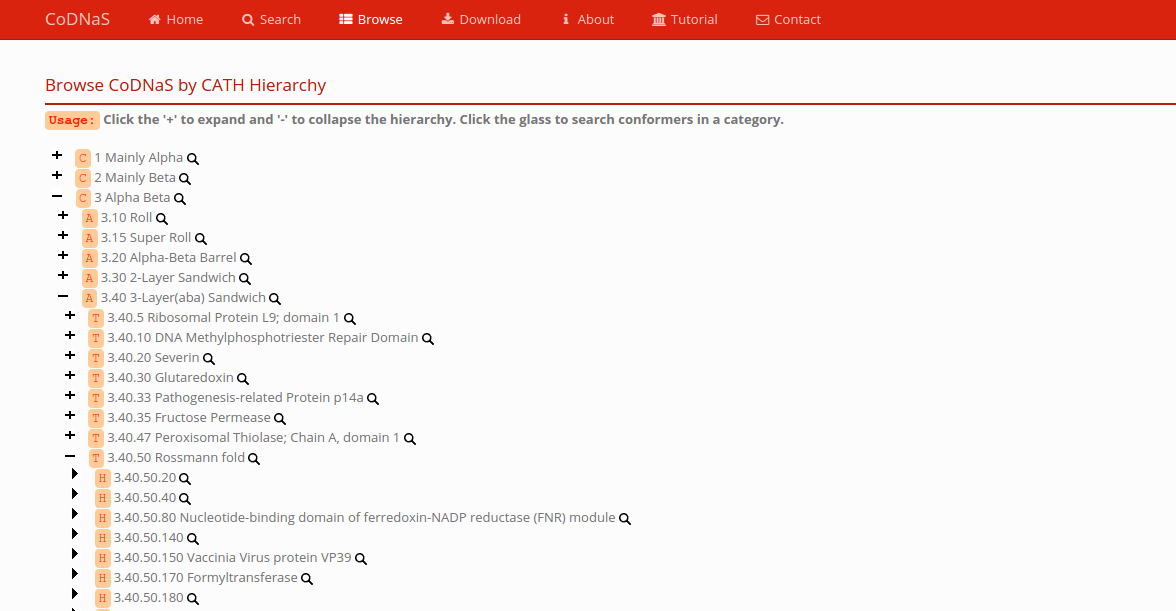
Browse
You can browse CoDNaS according to the CATH hierarchy. Expand the tree to show the category of interest, click the glass, and you will get all the conformers that belongs to this category.